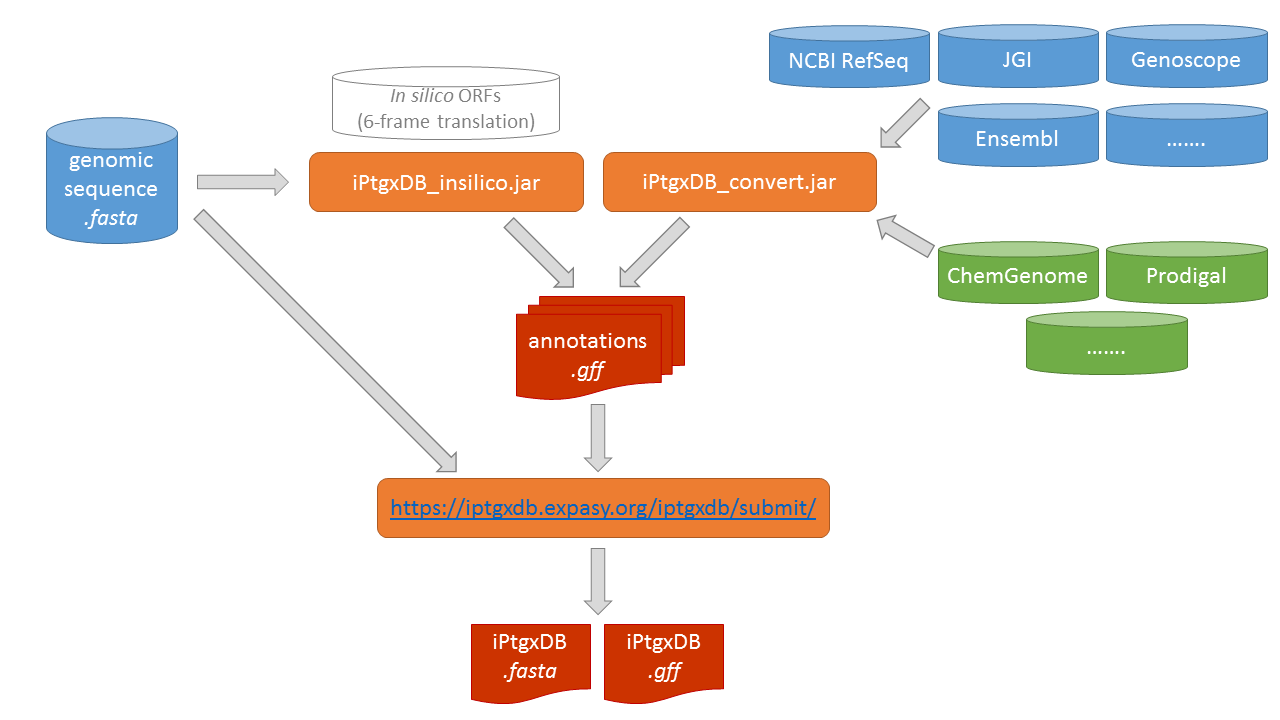
We release the code to generate custom iPtgxDBs in the form of two java jar files and a public web server. Using a genome sequence file in FASTA format as
input, the script iPtgxDB_insilico.jar
can be executed to create a 6-frame translation (with additional options like alternative start codons). One or
several existing reference genome annotations (blue containers) or results of ab initio gene prediction
tools (green containers) can be processed with iPtgxDB_convert.jar to create
several annotation.gff files (one per input file). All of these gff files are then combined on our public web server to create both the searchable iPtgxDB.fasta file
and the iPtgxDB.gff file that contains all integrated annotations.